Challenge & Context
As a consequence of the rapid acceleration of digital transformation, cities are gradually starting to publicly release urban data. However, most data is currently released in formats that are not processable. Even though most businesses and public administrations are rich in data, few have mastered the art of using it in an open and secure manner to analyze what is happening in their organization and cities and, hence, create opportunities for digital transformation and ultimately enhance citizens’ lives.
Now is the time for change. Engineering has joined the battle against the coronavirus epidemic SARS-CoV-2 (hereinafter referred to as “COVID-19”) with its ready-to-use FIWARE-based biosurveillance system that helps public administrations and health institutions to have a ‘near’ real-time holistic view of contextualized information coming from different data sources. This way, public administrations and health institutions can better understand their priorities, define and apply epidemiological models, and real-time geolocation of some clinical-health phenomena, and predictive maps of contagion. The solution was developed for the Veneto region, in Northeast Italy, which has been heavily affected by COVID-19.
The evolution of COVID-19, characterized by a high index of infection and rapid spreading, makes (‘near’) real-time monitoring of the evolution of the pandemic even more indispensable. Speeding up the identification, isolation and monitoring of infected people is therefore an important factor in the fight against COVID-19, and Engineering, which has, for decades, been developing innovative digital solutions, has delivered a cloud-based solution to aid this process. By relying on the many features of Engineering’s Digital Enabler, a ‘Powered by FIWARE’ solution that not only identifies potential data sources within a given ecosystem but also collects insightful data and eases its contextualized visualization and usage through intuitive and replicable dashboards, Engineering has developed the Eng-DE4Bios platform.
In the context of the spread of an pandemic such as that of COVID-19, in which the growth curve has an exponential trend, some of the features of the Eng-DE4Bios platform, such as the flexibility and simplicity to switch from the mock-up of a service to its actual publication, become even more important, reducing time to market and improving decision making responsiveness over monitored data.
Lorenzo Gubian, CIO Healthcare Department at Veneto region, describes how “the biosurveillance system developed by Engineering allowed us to monitor in real-time the pandemic spread, and provided us with data to predict its effects in advance. All this has allowed us to put in place preventive actions to govern the emergency system, avoiding further infections, containing the number of people who die and mainly saving lives”
The first 3 positive cases in Veneto were detected on February 21. The idea to create a biosurveillance system for COVID-19 was born the same night. Engineering started started working on the Digital Enabler on February 26, and after 7 days the first service for general practitioners started. From March 8 the platform was up and running.
Arianna Cocchiglia, Director of Health Innovation at Engineering confirms that “the flexibility in integrations and the simplicity to integrate different data sources have allowed us to have a real-time data collection system in a very short time, this is not normally the case in healthcare solutions”
Solution
The biosurveillance system, Eng-DE4Bios, currently in use in the Veneto region, is based on the Digital Enabler platform and relies on various of its features. The Digital Enabler is a data-driven, native cloud, ecosystem platform designed by Engineering’s R&D Labs. Today, it is one of the few fully functioning, ready-to-use, cloud ecosystem platforms available on the market.
As an “ecosystem” platform, the Digital Enabler empowers new data economy business models to flourish, fostering innovation and enhancing existing capabilities. Thanks to FIWARE Open Source technology, solutions can be built to address emergency situations in a short amount of time. These solutions are easily replicable and can be adopted and used by different regions and nations.
The Digital Enabler should be used wherever mission-critical information needs to be enriched, in order to take fast and educated decisions:
- to create an open innovation area, to manage and develop new ideas;
- to discover and harmonise different types of data sources (Open Data, Enterprise Data, IoT devices, and infrastructure, to name but a few) increasing users’ base knowledge;
- to evaluate the quality of the data sources, improving overall data quality;
- to provide real-time data management and event-driven information to alert and monitor;
- to dynamically model and integrate data;
- to visualize, analyze and render data via mash-up and mock-up capabilities and frontend dashboards, creating new digital capabilities, applications, and services to power up the new data economy ecosystem.
The Digital Enabler allows users to harmonize, synchronize, integrate, visualize, mashup, federate, and analyze data to deliver their digital transformation vision and urgent requirements. In addition, it provides a single point of data knowledge and access that can be used to develop new value-added services as well as digitally enabling older technologies and applications:
- Data discovery: identify potential data sources within an ecosystem.
- Data collection: collect insightful data and make it easily exploitable via the platform.
- Data integration: mash-up multi-source data to create new knowledge.
- Data harmonization: harmonize data to FIWARE standard data models.
- Data visualization: visualize data through intuitive and replicable dashboards.
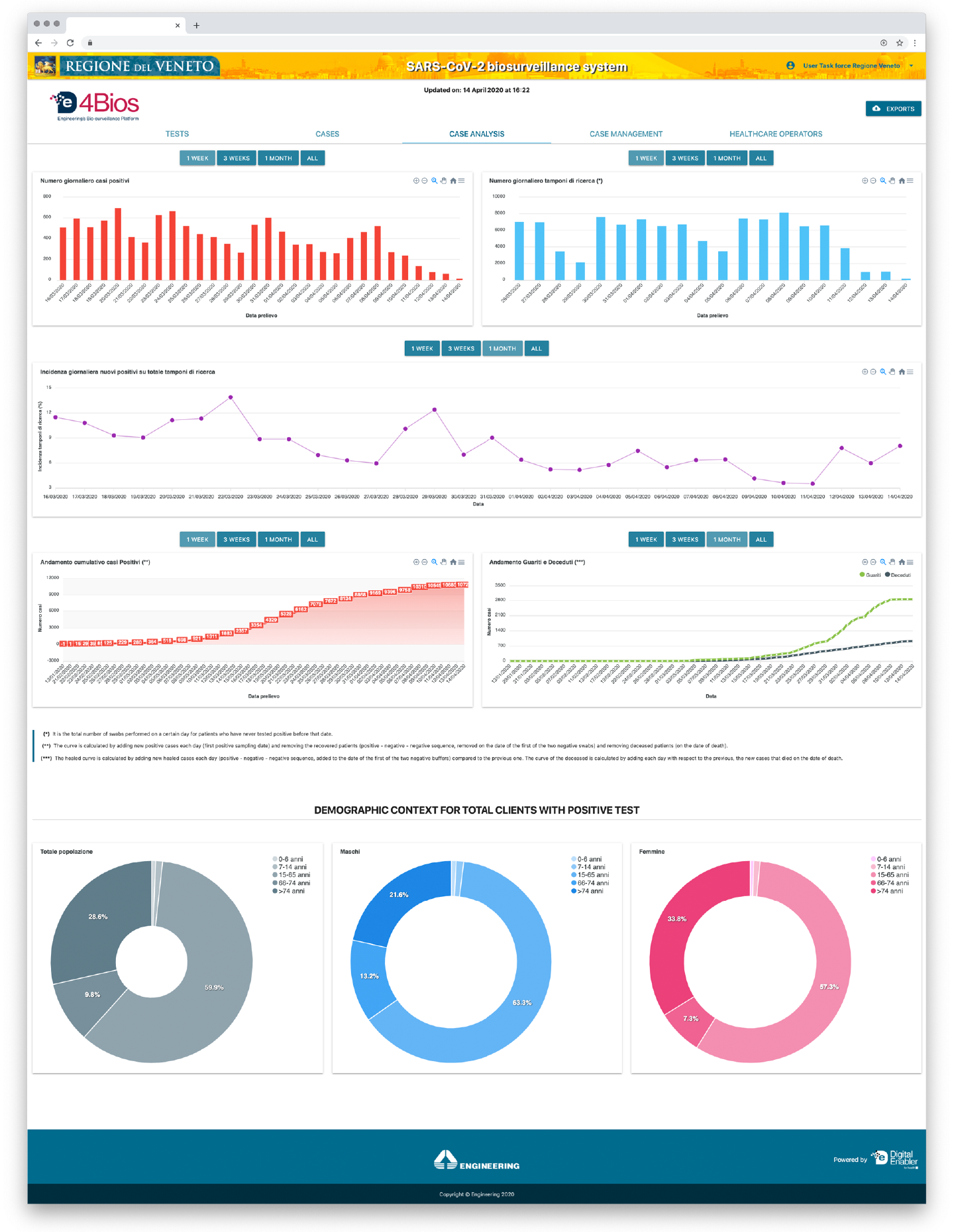
Figure 1 . De4Bios Analytics
How it works
Biosurveillance is an area related to the medical sector in which the detection of and resistance to pathogens is monitored regularly and information is collected by a central system for the analysis and understanding of the epidemic phenomenon.
Biosurveillance is the process of collecting, integrating, interpreting and communicating the essential information relating to threats, dangers or disease activity affecting human, animal or plant health to get early observations and warnings. It supports a clear insight into the overall situation of the clinical-health aspects and enables a better decision-making process at all levels. Compared to previous years, biosurveillance systems now need to deliver results faster, at ‘near’ real-time speed, to help the overwhelmed medical system to understand priorities and quickly take tangible, high-impact measures.
Eng-DE4Bios supports public administrations to manage epidemic crisis by providing:
- to medical doctors: a real-time view of their assisted population who performed the COVID-19 swab with the result of the test and a quick geo-localization on a map of the positive cases.
- to COVID-19 crisis units: a real-time updated view of all monitoring data, accordingly to WHO and HL7 standards:
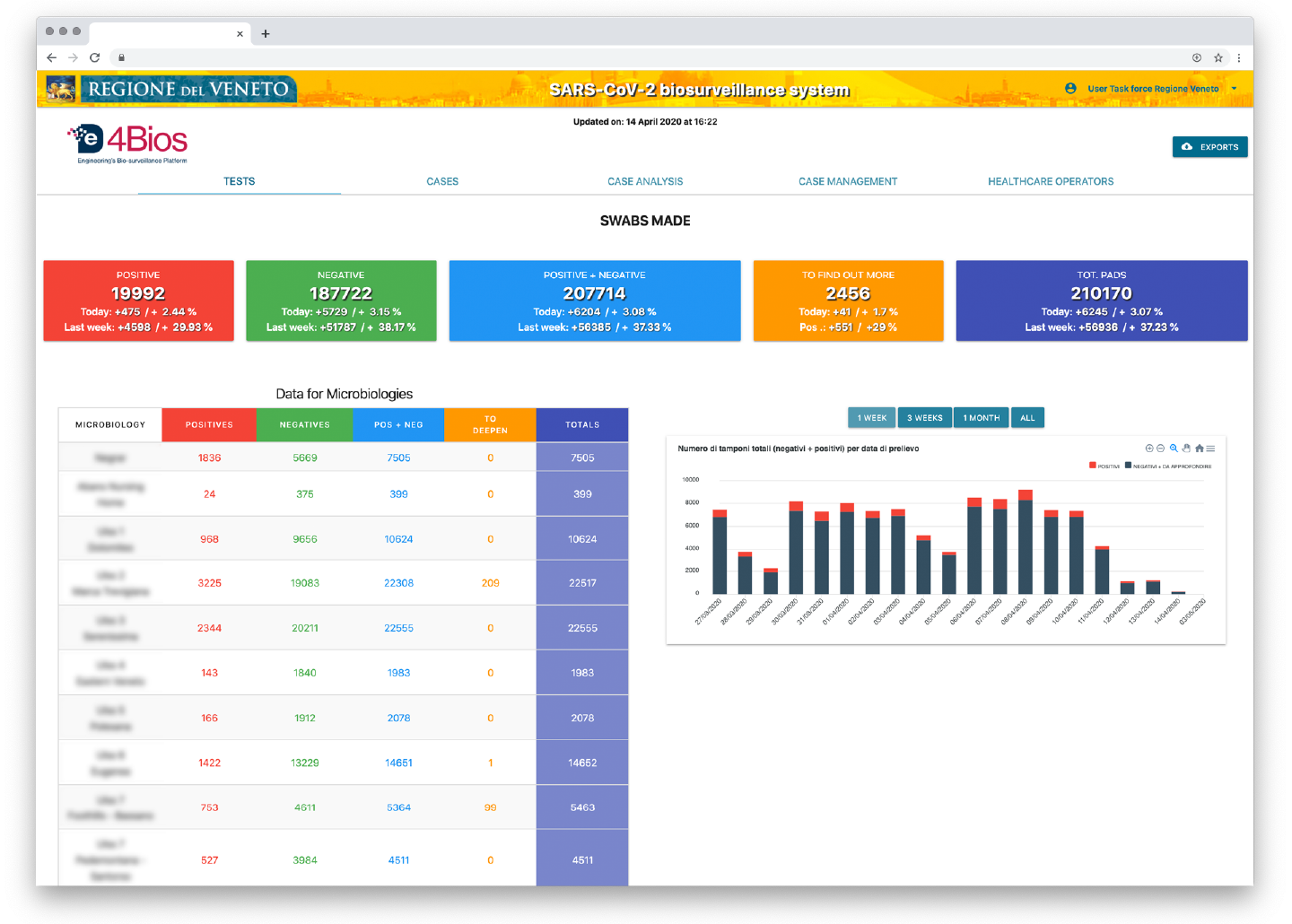
- aggregated data on the results of the swabs carried out (positive, negative, to be further investigated, or total) with details related to each single microbiology lab that performed the tests;
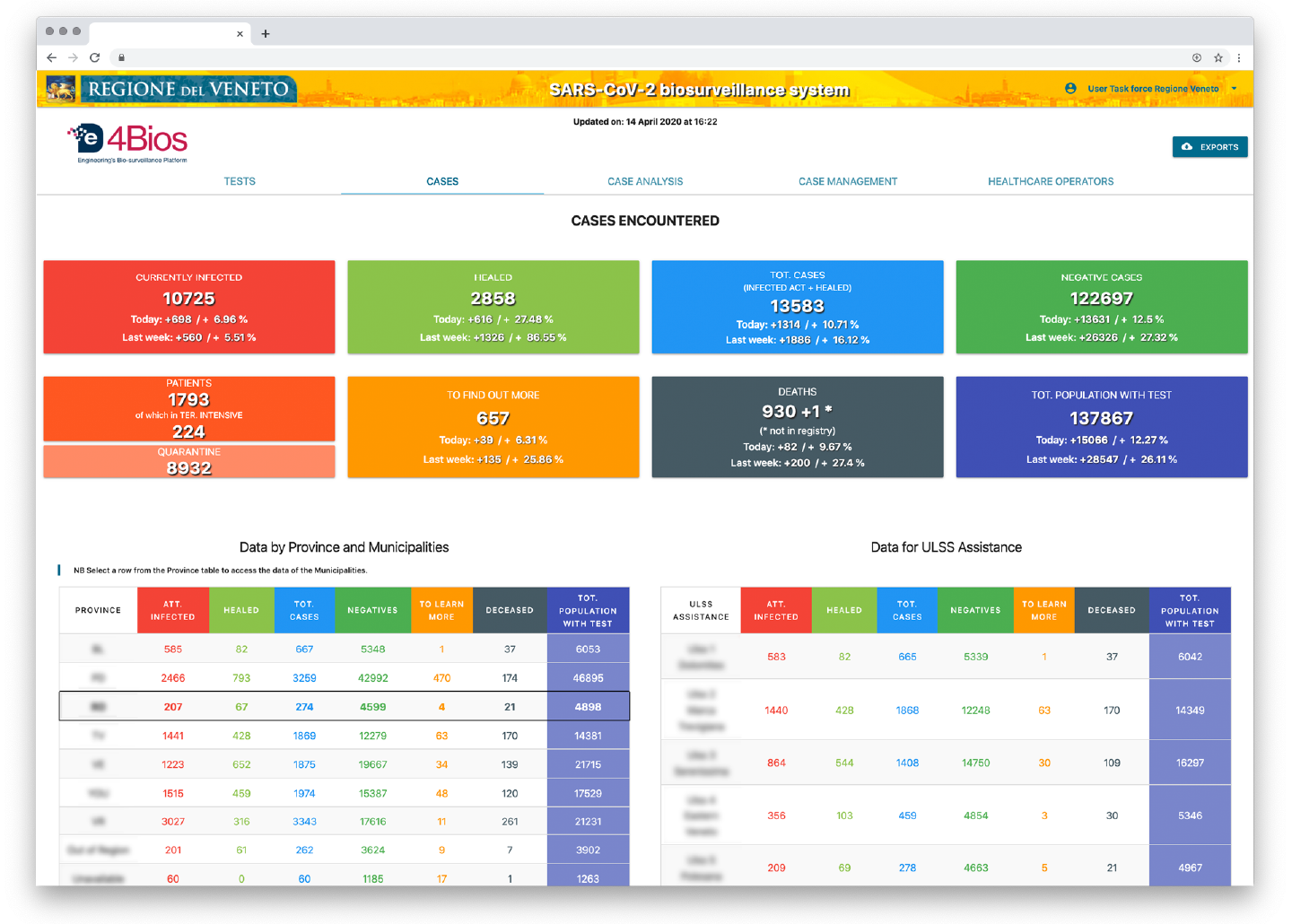
- aggregated data on cases, i.e. people, divided by ‘currently positive’ (people who have at least one positive swab), ‘healed’ (people who have one positive swab and two consecutive negatives), ‘negative’ (people who have only one or more negative swab), ‘total population’ (people who undergo at least one swab). This aggregated information is also made available to municipalities and to healthcare organizations;
- the demographic context (e.g. age groups, population under screening, or infected population);
- evolution curves of the epidemic (infected, infected, or healed);
- correlated database of the population with the possibility of multi-queries sorted by test results, doctor, healthcare authorities, municipality, test date, test report date, etc.;
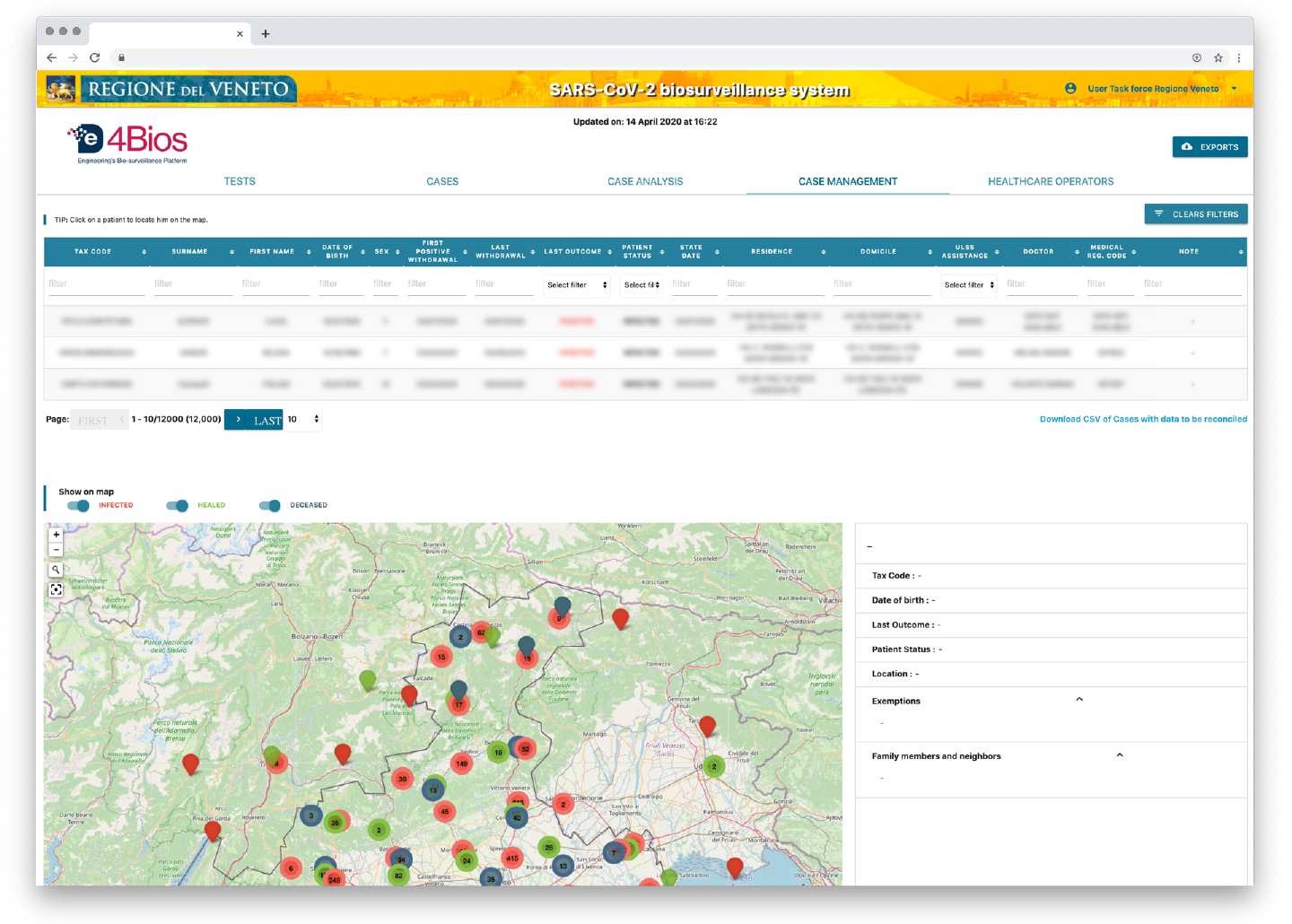
- positive cases geo-referenced on a map aggregated by territorial incidence, navigable up to the identification of each individual case.
- to crisis units of each individual health authority: the same view described above, but only with the data of the patients related to it.
The Eng-DE4Bios biosurveillance system combines very heterogeneous data sources to create databases on which to perform Advanced Data Analytics and to define and apply epidemiological models, real-time geolocation of some clinical-health phenomena, and predictive maps of contagion.
Figure 2 . De4Bios Maps
Benefits & Impact
Eng-DE4Bios delivers:
- real-time information on the status of the infected population (infected, healed, deceased, hospitalized, or hospitalized in intensive care);
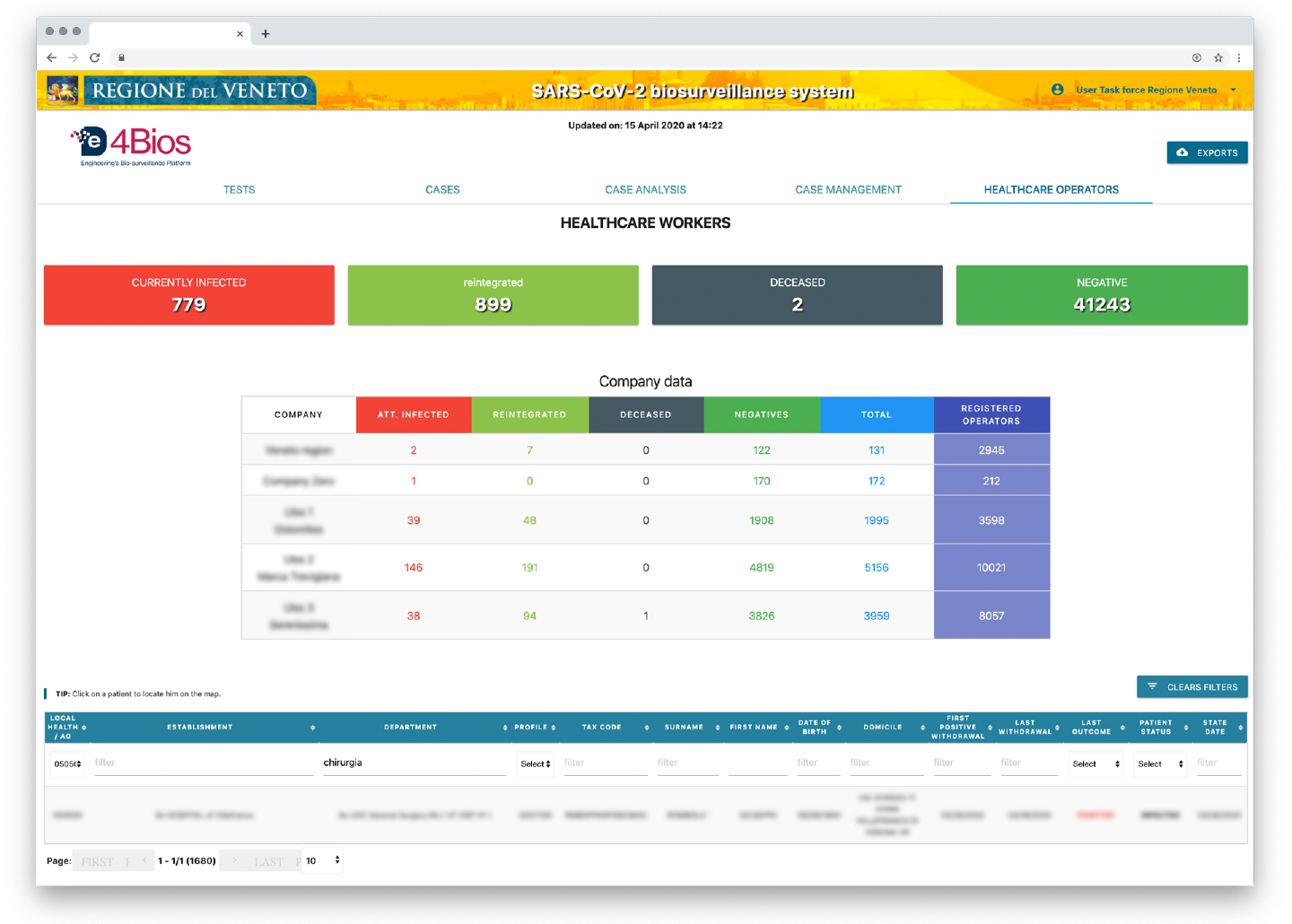
- monitoring of the virus spread among health workers and their operations (infected and isolated workers), in order to evaluate the decrease or increase in operational forces;
- correlation services which, starting from a positive case, identify other probable people exposed to infection due to contact with other members of the same family group and/or living at the same address, it also identifies which of those have already carried out the swab (with the related result) and which have not yet carried out the swab (identifying people most at risk by age group, pathologies, etc.);
- a service that, starting from a positive case, identifies if the infected person is an employer/student and other related information (work address/school address) in order to reconstruct the network of possible infected people.
The systematic collection of all the above information allows public administrations and health institutions to have at their disposal a database to implement different types of epidemiological models and represent the best possible evolution of the epidemic. These models can be implemented as an application that allows the automatic simulation and calibration of the epidemic, with the possibility to forecast scenarios when the parameters of the model change.
Figure 3 . De4Bios Tests
Figure 4 . De4Bios Cases
Figure 5 . De4Bios Caregivers
Added value through FIWARE
The solution is entirely based on open standards and open APIs, and data models, according to FIWARE Foundation and OASC principles (https://oascities.org/). It is ‘Powered by FIWARE’, using FIWARE Generic Enablers. By using FIWARE NGSI APIs (https://fiware.github.io/specifications/ngsiv2/stable/), which easily enable the integration of different components and systems, Engineering ensures that the platform is interoperable, portable, and can be easily implemented in other neighbouring regions and nations, as well as on a global scale. As the Digital Enabler is part of an already existing solution helping to mitigate the spread of the virus, the gains here are of unparalleled magnitude. Not only can this cloud-based platform be connected with other applications or pieces of software already widely available, but it can also be replicated to serve various other purposes at a fast speed and lower costs.
At the center of the system lies the Context Broker, a FIWARE Generic Enabler and a Connecting Europe Facility (CEF) Building Block. CEF is an EU funding program that promotes growth and competitiveness through targeted infrastructure investment at a European level. The CEF Context Broker is an API that can integrate data from multiple systems, creating a holistic view of contextualized information. By providing the layer that describes each type of data, the Context Broker makes it possible to create an interface that makes it easy for anyone to view and interpret big data.
The CEF Context Broker is built using FIWARE NGSI specifications that enable applications to provide real-time updates and access to information that contextualizes the data being displayed. Typical examples might include updates, queries, and notifications. This way, organizations can monitor their metrics in real-time through live updates. Context information can be shared with third-parties, enabling process improvements and innovation across the whole data value chain.
Next Steps
Accessing data sources to be integrated in the first phase of the services include:
- test results from microbiology labs enabled to perform the screening for COVID-19. The system integrates the personal data of the patients who were screened and the relative outcome;
- regional/national health registries to reconcile the records from the results of the microbiologists with a certain registry position and expand the database with information regarding missing or incomplete personal data or healthcare data;
- registry to obtain data of the nucleus of the patients who tested positive;
- data on the status of the infected population (infected, healed, deceased, hospitalized, or hospitalized in intensive care). In more detail, the infected and the healed population are identified automatically by analyzing the results of the tests while the number of people deceased, hospitalized and hospitalized in intensive care can be made available to the system through an ad-hoc integration or through the implementation of a web application;
- administrative flows from hospitals and first aid;
- regional worker registries or social security databases from which the data of the employer, headquarters address and contacts for positive people can be retrieved;
- Personal Health Records (PHR) to have real-time clinical data and documents and any non-patient-centric information.